竞赛圈 > 菜鸟解法。kmeans+线性回归
我这解法分数很低的。但之前用这方法是0.06多。今天重新实现了一下变成了0.13多了。。。时间仓促也是一部分原因,没有调就交了。至少是个思路。。。
我使用R语言。
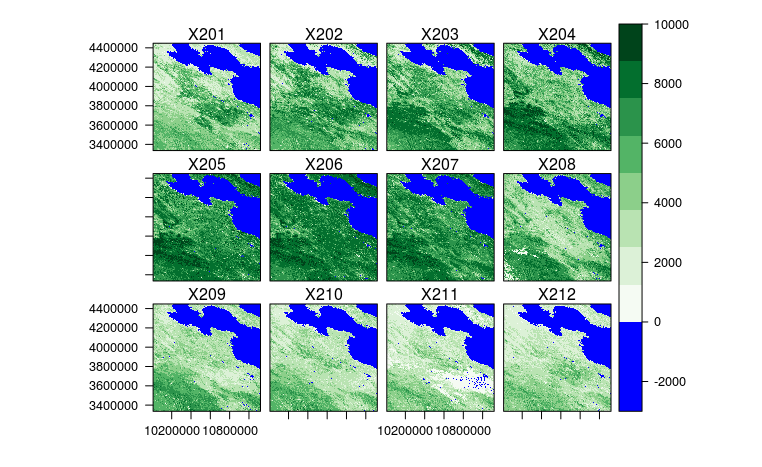
探索性数据分析:我使用raster包读数据,rasterVis包画图。也不说太多,也就说一点点发现。首先,第三个区域有大片海洋。其次,有些图片有明显的云彩,有的可能对预测会产生很大的影响,例如第三个区域的第211个月,图片中下部分明显有云,而如果不考虑这个的话,肯定会对213个月的预测产生很大的影响(可惜我算法里还没能实现处理这种异常值的方法)。第三,相邻的像素很可能是不连续,而是有突变的。第四,好像从图中隐约可以看出南方的春天来的比北方早,不确定。。。下图是第3个区域的最后12个月。
算法:最开始我是直接逐个像素跑线性回归。由于数据量比较大,稍微fancy一点的统计方法根本跑不动。。。由于数据量比较大,直接跑回归也得跑自己手动实现的线性回归,直接上lm可能要跑几天。自己实现的线性回归,整个跑下来大概40分钟吧。这个其实是我的最高分,0.060多一点。
然后这样至少有三个问题。第一,其实对于逐像素点回归,数据量并不大,甚至是不够的。第二,对于一个像素点的预测,没有利用其他点的信息,可以想见,其实其他点也一定是包含信息的。第三,从残差可以看出,明显有一些异常点,猜测应该是云。
对第二个问题,我有一点想法,但还没来得及实现。。对于第三个的话,也许神经网络有可能利用其他点的信息,不确定。我主要尝试了对一个问题的解决。首先想做的是Mixed model,用lme4包。这样可以把所有数据放一起训练个大模型。可惜数据量过大,计算起来根本不可行,即使分成很小的块也是不行的。下面的代码是我的另一个尝试,也就是先用kmeans对像素点进行聚类(聚成5类),每类里聚合成大数据集,然后跑回归,这样跑回归就不是速度瓶颈,可以直接lm。速度瓶颈在于构造训练集,内存瓶颈也在此,把训练集构造的大一点就很容易爆掉内存。这样做是可行的,得到的模型也很显著,但我还没调好,协变量没调好,这可能是分数较低的原因。我觉得调整良好的这个模型可以比逐点线性回归效果好,后面也可能对异常值的处理更加。
下面是代码,任何人都可以用作任何用途。
1. preProcess.R 数据预处理,把1200*1200的区域分成9个400*400的区域并保存
library(raster)
#library(Rfast)
for (theFolder in c("Z1","Z2","Z3","Z4")){
gc()
# obtain numbers of files
theNumbers <- length(dir(paste0("../data/",theFolder)))
# generate all file names
myDataList <- list()
myDataValues <- list()
tmpNum <- as.character(1:theNumbers)
tmpNum <- paste0("000",tmpNum)
tmpNum <- substr(tmpNum,nchar(tmpNum)-2,nchar(tmpNum))
myFilenames <- paste0(theFolder,"-",tmpNum,".tif")
# read all data
for(tmpName in myFilenames){
myDataList[[tmpName]] <- raster(paste0("../data/",theFolder,"/",tmpName))
}
names(myDataList)<- 1:length(myDataList)
myDataValues <- array(dim=c(length(myDataList),nrow(myDataList[[1]]),ncol(myDataList[[1]])))
for(i in 1:dim(myDataValues)[1]){
myDataValues[i,,] <- as.matrix(myDataList[[i]])
}
tmpNrow <- floor(dim(myDataValues)[2]/3)
tmpNcol <- floor(dim(myDataValues)[3]/3)
for(rN in 1:3)for(cN in 1:3){
saveRDS(myDataValues[,(tmpNrow*(rN-1)+1):(tmpNrow*rN),(tmpNrow*(cN-1)+1):(tmpNrow*cN)],
paste0("./tmpData/",theFolder,"-data-",rN,cN,".rds")
)
}
}
gc()
2. enter.R 入口,对每个小区域调用模型
for(theFolder in c("Z1","Z2","Z3","Z4"))for(rN in 1:3)for(cN in 1:3){
source("./model.R")
}3. model.R 模型
#library(raster)
#library(TSA)
#library(Rfast)
library(parallel)
#library(dplyr)
#library(compiler)
#enableJIT(1)
gc()
myDataValues <- readRDS(paste0("./tmpData/",theFolder,"-data-",rN,cN,".rds"))
myLength <- dim(myDataValues)[1]
gc()
# means
theJiuMean <- apply(myDataValues,c(2,3), mean)
# Clustering
myDataValuesTemp <- myDataValues
dim(myDataValuesTemp) <- c(myLength, 400^2)
myDataValuesTemp <- t(myDataValuesTemp)
theCluster <- kmeans(myDataValuesTemp,centers = 5)$cluster
dim(theCluster) <- c(400,400)
rm(myDataValuesTemp)
gc()
# Add cluster to Data
dim(theCluster) <- c(1,400^2)
dim(myDataValues) <- c(myLength,400^2)
myDataValues <- rbind(theCluster,myDataValues)
dim(theCluster) <- c(400,400)
dim(myDataValues) <- c(myLength+1,400,400)
# construct training Data
youyou <- c(1,2,12,24)
myConstructFunction <- function(myTS){
ohCluster <- myTS[1]
myTS <- myTS[-1]
myIndex <- 1:length(myTS)
youyou <- c(1,2,12,24)
myIndexList <- lapply(youyou,function(i){myIndex - i})
names(myIndexList) <- youyou
tmp <- myIndexList[[as.character(youyou[length(youyou)])]] > 0
# these two lines are new
tmp2 <- myIndex %%12 == (length(myTS)+1) %%12
tmp <-tmp & tmp2
y <- myTS[myIndex[tmp]]
tmpx <- lapply(myIndexList,function(zzz){myTS[zzz[tmp]]})
names(tmpx) <- names(myIndexList)
x <- cbind(do.call(cbind,tmpx) )
data.frame(y,x,myMean = mean(myTS), ohCluster)
}
cl <- makeCluster(3)
clusterExport(cl,c("myLength"))
#clusterEvalQ(cl,library("Rfast"))
myHahaData <- do.call(rbind,parApply(cl,myDataValues,c(2,3),myConstructFunction))
stopCluster(cl)
gc()
myHahaData2 <- split(myHahaData,myHahaData$ohCluster)
myFormula <- as.formula(paste0("y~",paste(paste0("X",youyou),collapse=" + "),"+","myMean"))
myModels <- lapply(myHahaData2,function(data){
tmp <- lm(myFormula,data)
tmp2 <- coef(tmp)
tmp2[is.na(tmp2)] <- 0
tmp2
})
#myModel <- lm(y~.-1,myHahaData)
#rm(myHahaData)
#myModel$model<- NULL
#gc()
myDataValues <- myDataValues[(myLength-30):myLength,,]
gc()
# Predict
myPredict<- rep(0,400^2*3)
dim(myPredict) <- c(3,400,400)
for(n1 in 1:400)for(n2 in 1:400){
y <- myDataValues[,n1,n2]
# testing
#tmp$meanInterval <- cut(theJiuMean[n1,n2],myBreak)
for(i in 1:3){
#tmp[1,2:6] <- c(y[length(y)+1-youyou],theJiuMean[n1,n2])
tmp <- c(1,y[length(y)+1-youyou],theJiuMean[n1,n2])
y[length(y)+1] <- sum(myModels[[theCluster[n1,n2]]]*tmp)
}
myPredict[,n1,n2] <- y[length(y)-3+1:3]
}
# myDistributedFunction <- function(myTS){
# myOut <- rep(0,3)
# # model 1
# #yuShu <- (length(myTS)+1)%%12
# #myIndex <- which((1:length(myTS))%%12==yuShu )
# myIndex <- 1:length(myTS)
# y <- myTS
# youyou <- c(1,2,12,24)
# # testing
# for(i in 1:3){
# tmp <- c(1,y[length(y)+1-youyou])
# y[length(y)+1] <- sum( c(1,y[length(y)+1-youyou]) * be )
# }
#
# myOut <- y[length(y)-3+1:3]
# myOut
# }
#
# myPredict <- parApply(cl,myDataValues,c(2,3),myDistributedFunction)
saveRDS(myPredict,paste0("./tmpData/",theFolder,"-output-",rN,cN,".rds"))4. outPut.R 输出
library(raster)
# ooTmp <- raster(myDataList[[1]])
# values(ooTmp) <- 0
# tmp <- list()
# tmp[[1]] <- ooTmp
# tmp[[2]] <- ooTmp
# tmp[[3]] <- ooTmp
#
# for(i in 1:3){
# tmp[[i]][myRowNumber,myColNumber] <- myTestingY[i]
# }
#
# for(i in 1:3){
# writename <- paste0(theFolder,"-",i,".tif")
# writeRaster(my12Sums[[tmp]],paste0("../results/",writename))
# }
theFolder <- "Z4"
theNumbers <- length(dir(paste0("../data/",theFolder)))
myRowList <- list()
for(rN in 1:3){
tmpList <- list()
for(cN in 1:3){
tmpList[[cN]] <- readRDS(paste0("./tmpData/",theFolder,"-output-",rN,cN,".rds"))
}
myRowList[[rN]] <- tmpList
}
myList2 <- list()
for(i in 1:3){
tmpList <- lapply(myRowList,function(tiantian){
momo <- lapply(tiantian,function(data){data[i,,]})
do.call(cbind,momo)
})
#myList2[[i]] <- cbind(rbind(myList[['11']][i,,],myList[['21']][i,,]), rbind(myList[['12']][i,,],myList[['22']][i,,]))
myList2[[i]] <- do.call(rbind,tmpList)
}
for(i in 1:3){
writename <- paste0(theFolder,"-",i+theNumbers,".tif")
writeRaster(raster(myList2[[i]]),paste0("../results/",writename))
}
5. explanatoryDataAnalysis.R 画图
# heat map
# library(ggplot2)
#library(lattice)
library(rasterVis)
myTheme <- rasterTheme(region = c("#0000FF",brewer.pal("Greens",n=9)))
myBigData <- stack(myDataList[201:212])
levelplot(myBigData,at=c(-3e3,seq(0,1e4,length.out=9)),par.settings=myTheme)
# results
# training data
theFolder <- "Z3"
theNumbers <- length(dir(paste0("../data/",theFolder)))
# generate all file names
myDataList <- list()
myDataValues <- list()
tmpNum <- as.character(1:theNumbers)
tmpNum <- paste0("000",tmpNum)
tmpNum <- substr(tmpNum,nchar(tmpNum)-2,nchar(tmpNum))
myFilenames <- paste0("../data/",theFolder,"/",theFolder,"-",tmpNum,".tif")
# read all data
for(tmpName in myFilenames){
myDataList[[tmpName]] <- raster(tmpName)
}
names(myDataList)<- 1:length(myDataList)
# results
tmpNum <- as.character((theNumbers+1):(theNumbers + 3))
tmpNum <- paste0("000",tmpNum)
tmpNum <- substr(tmpNum,nchar(tmpNum)-2,nchar(tmpNum))
myFilenames2 <- paste0("../results/","/",theFolder,"-",tmpNum,".tif")
myPredictList <- list()
for(tmpName in myFilenames2){
myPredictList[[tmpName]] <- raster(tmpName)
}
names(myPredictList)<- (1:length(myPredictList))+212
for(i in 1:3){
myPredictList[[i]]@extent <- myDataList[[1]]@extent
}
# levelplot
myTheme <- rasterTheme(region = c("#0000FF",brewer.pal("Greens",n=9)))
myBigData <- stack(c(myDataList[204:212],myPredictList[1:3]))
#myBigData <- stack(myPredictList[1:3])
levelplot(myBigData,at=c(-3e3,seq(0,1e4,length.out=9)),par.settings=myTheme)
 关注微信公众号
关注微信公众号